The bacterium displaced a fluorine atom attached to the double bond of a specific type of PFAS known as PFMeUPA but did not remove fluorine atoms attached to saturated carbon atoms.
One of the key characteristics that enabled this bacterium to partially decompose PFMeUPA is the presence of a channel that facilitates removal of fluoride ions from the bacterium cell and is known as a fluoride anion exporter.
PFAS, per- and polyfluorinated alkyl substances, are under regulatory scrutiny due to health and safety concerns. Due to their stability, they are designated as “forever chemicals” making PFAS difficult to identify and to decompose, rendering them harmless in the environment.
A previous TLT article
1 focused on the key health and safety concerns in dealing with PFAS. There are now approximately 15,000 PFAS in existence according to the U.S. Environmental Protection Agency (EPA) making it difficult to determine the toxicity of each individual substance. As pointed out in this article, methods for isolating PFAS are much more prevalent than strategies for identifying and destroying them. This is leading to an imbalance making it very challenging to manage PFAS contamination in the environment. The advice given is for organizations working with lubricants to devise a strategy to determine what sources are most likely culprits containing PFAS.
Yujie Men, associate professor in the Bourns College of Engineering in the Department of Chemical and Environmental Engineering at the University of California, Riverside in Riverside, Calif., says, “The difficulties faced in decomposing PFAS are due to the strength of carbon-fluorine bonds, which are difficult to break. Compared to physiochemical approaches, it would still be ideal to remediate PFAS-impacted sites with microbes, which are more suitable for in situ remediation and environmentally friendly, if microbes that contain effective defluorinating enzymes could be discovered.”
Water is one of the main sources of PFAS contamination. As shown in Figure 1, bacteria are readily used to treat groundwater in a cost-effective manner. If present in sufficient quantity, bacteria can consume pollutants that originate from a point source such as a factory.
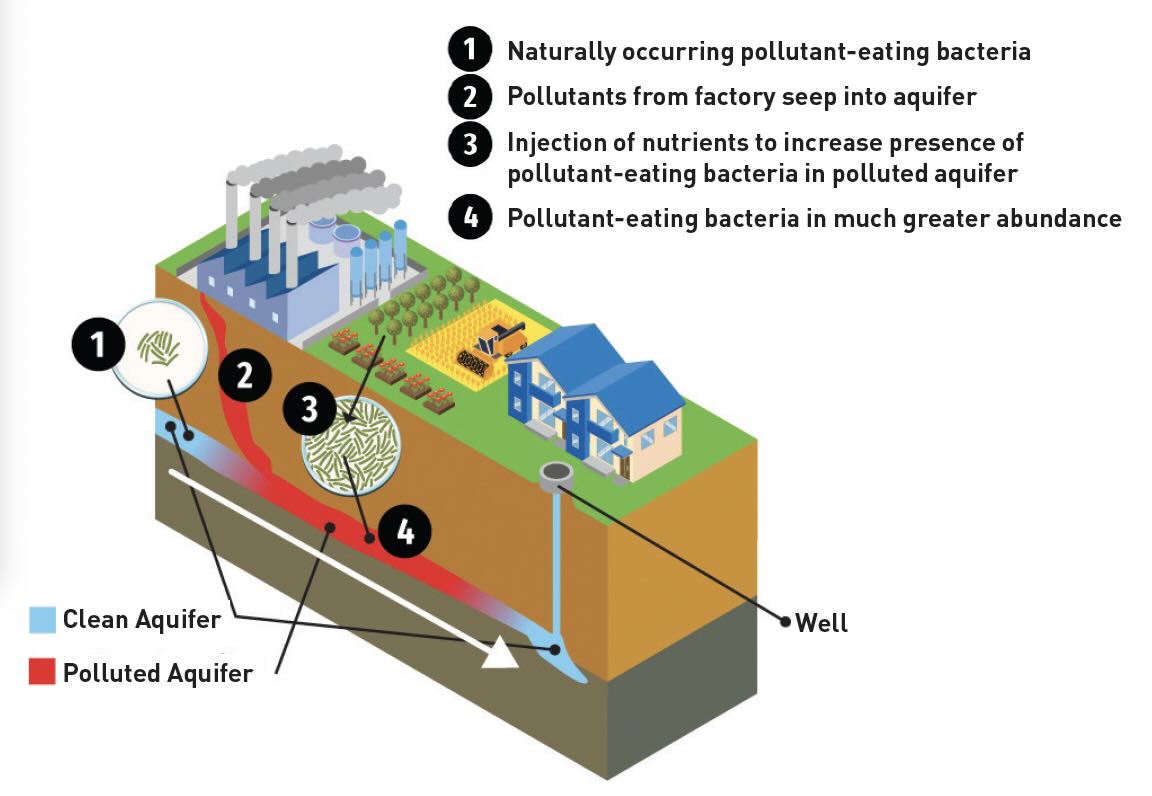 Figure 1. Bacteria appear to be a viable option for decomposing PFAS present in water because they are widely used in waste treatment of ground water. Figure courtesy of the University of California, Riverside.
Figure 1. Bacteria appear to be a viable option for decomposing PFAS present in water because they are widely used in waste treatment of ground water. Figure courtesy of the University of California, Riverside.
Men says, “The potential approach to decompose PFAS is to use enzymes contained in specific bacteria in a reductive defluorination process where fluorine will be replaced with hydrogen atoms. While this process is thermodynamically favorable, identifying the right enzymes to achieve this object is difficult because of the high energy required, concern about fluoride toxicity toward bacteria and the short evolutionary time span where bacteria are exposed to PFAS.”
Another limitation is that no bacterium or enzyme in common environments had been found that will reductively defluorinate fully fluorinated structures.
A potential strategy for dealing with PFAS originated from past work conducted by Men and her colleagues. She says, “We found that exposure of a specific, unsaturated PFAS known as PFMeUPA (E-perfluoro-4-methylpent-2-enoic acid) to an anerobic bacteria community led to defluorination though did not identify the specific bacterium or enzyme involved.”
The next step was to work with PFMeUPA and determine which bacteria are directly involved in reductive biodefluorination.
Fluoride anion exporter
Bacteria used in the study were grown in the incubator shown in Figure 2.
 Figure 2. Bacteria that were used to evaluate PFAS were grown in an incubator. Figure courtesy of the University of California, Riverside.
Figure 2. Bacteria that were used to evaluate PFAS were grown in an incubator. Figure courtesy of the University of California, Riverside.
Initially, the researchers conducted a screening test to determine if two pure bacteria species displayed any evidence of defluorination. They found that
Acetobacterium bakii defluorinated PFMeUPA within three weeks. After conducting mass spectroscopy and
19F nuclear magnetic spectroscopy, the researchers identified two microbial reductive defluorination transformation products.
Men says, “Our analysis showed that the bacterium displaced a fluorine atom attached to the double bond of PFMeUPA and replaced it with a hydrogen atom. Hydrogenation of PFMeUPA reduced the double bond to a single bond yielding the second product. PFMeUPA only removed one fluorine atom leaving the remaining eight fluorine atoms untouched. This includes fluorine atoms bonded to saturated carbon atoms.”
Two other PFAS with similar unsaturated structures to PFMeUPA also were reductively defluorinated in a similar manner by
Acetobacterium bakii.
Further testing indicated that no defluorination occurred when PFMeUPA is exposed to a cell-free medium. This means that enzymes within the bacterium cell must be carrying out defluorination.
Four additional
Acetobacterium species were then evaluated for defluorination activity. The researchers determined that all but one could defluorinated PFMeUPA. The one that was not able to remove a fluorine atom from the unsaturated PFAS did not contain a functional fluoride anion exporter.
Men says, “A fluoride anion exporter is a channel in the membrane of a bacterium that facilitates the removal of fluoride ions from the cell. Without this ability, toxic fluoride ions accumulate in the cell leading to the cessation of activity. We suspect the reductive defluorination is taking place in the inner membrane of the bacterium cell.”
Present within the
Acetobacterium are enzymes that have been found to reduce non-fluorinated unsaturated carboxylic acids similar to PFMeUPA. The researchers speculate that the enzymes which can reduce caffeate also are responsible for reductive defluorination of PFMeUPA. Men says, “We found that when caffeate and PFMeUPA are present, no defluorination occurs until the former is depleted.”
Men says, “Our results demonstrate that certain types of PFAS can be decomposed by specific enzymes. We hope that our approach can be combined with a second technique to facilitate decomposition of additional PFAS.”
Future work will focus on gaining a better understanding of the mechanism for how specific unsaturated PFAS such as PFMeUPA are decomposed. Men says, “We believe further research will lead to the design of new or alteration of existing enzymes that may be able to decompose a broader range of PFAS. After all, current PFAS under regulatory scrutiny such as PFOA are completely saturated and cannot be decomposed by the bacteria used in this study.”
Additional information can be found in a recent article
2 or by contacting Men at
ymen@engr.ucr.edu.
REFERENCES
1.
Canter, N. (2024), “PFAS: Many questions, few answers,” TLT,
80 (9), pp. 14-20. Available at
www.stle.org/files/TLTArchives/2024/09_September/Tech_Beat.aspx.
2.
Yu, Y., Xu, F., Zhao, W., Thoma, C., Che, S., Richman, J., Jin, B., Zhu, Y., Xing, Y., Wackett, L and Men, Y. (2024), “Electron bifurcation and fluoride efflux systems implicated in defluorination of perfluorinated unsaturated carboxylic acids by
Acetobacterium spp,”
Science Advances, 10 (29), DOI:10.1126/sciadv.ado295.